Introduction to microbial etiology of periodontal diseases
The research on the microbiology of periodontal health and diseases has been the focus of intense investigation for several decades. Microbial biofilm in the gingival sulcus around teeth is the most important cause of periodontal diseases. It is a structured community of bacterial cells enclosed in a self-produced polymeric matrix 1. Although more than 700 different species are present in subgingival microbiota 2-4, only a few of these species are actually involved in the initiation and progression of the periodontal disease process. Moreover, research on the etiopathogenesis of periodontal diseases has suggested the role of environmental 5, behavioral 6, 7, and genetic 8 risk factors in periodontal disease progression, but most, if not all forms of periodontitis, should be viewed primarily as infectious diseases. Many technological advances have occurred in molecular techniques in the last few decades which have provided us the capability of performing high-throughput analysis of a large number of samples, circumventing some of the limitations of cultural techniques. These techniques are especially useful in studying periodontal microbiology because there are many periodontal pathogens which are not cultivable and require molecular techniques to identify their presence in the bacterial sample. In the following discussion, we shall study the role of micro-organisms in the etiopathogenesis of periodontal diseases and details of those microorganisms which have been most commonly associated with periodontal disease progression.
Historical aspect
As already described in “Dental plaque”, the period from 1880 to 1930 is known as the ‘golden age of microbiology’ 9. Scientists identified four different groups of potential etiologic agents (amoebae, spirochetes, fusiforms, and streptococci) for various periodontal diseases using the techniques available at that time (wet mounts or stained smear microscopy). Researchers suggested the specific plaque hypothesis based on these findings. However, with the advancements in bacterial identification techniques, many other bacterial species were identified in dental plaque derived from periodontitis patients. Studies conducted between 1930 and 1970 failed to identify any specific microorganism as the etiologic agent of periodontal diseases which led to the proposal of non-specific plaque hypothesis, according to which gross accumulation of dental plaque would be necessary and sufficient to cause periodontitis.
Later on, with the advancements in the field of microbiology, immunology, and molecular biology, numerous studies concluded a putative pathogenic role of many bacteria, mainly Gram-negative species in the etiopathogenesis of periodontal diseases. These species include A. actinomycetemcomitans, Tannerella forsythia, Porphyromonas gingivalis, Prevotella intermedia, Campylobacter rectus, Fusobacterium nucleatum, and Treponema denticola. Virulence factors produced by these microorganisms have been identified and their role in periodontal destruction is well established. These findings led to the return of the theory of specificity in the microbial etiology of periodontal diseases. Presently, many bacterial species have been identified which contribute to the microbial etiology of periodontal tissue destruction. The concept of “polymicrobial dysbiosis” (read “Host microbial interactions”) is presently being investigated to explain the role of specific microorganisms in causing periodontal destruction. In the following sections, we shall discuss various micro-organisms associated periodontal health and disease.
Bacterial taxonomy
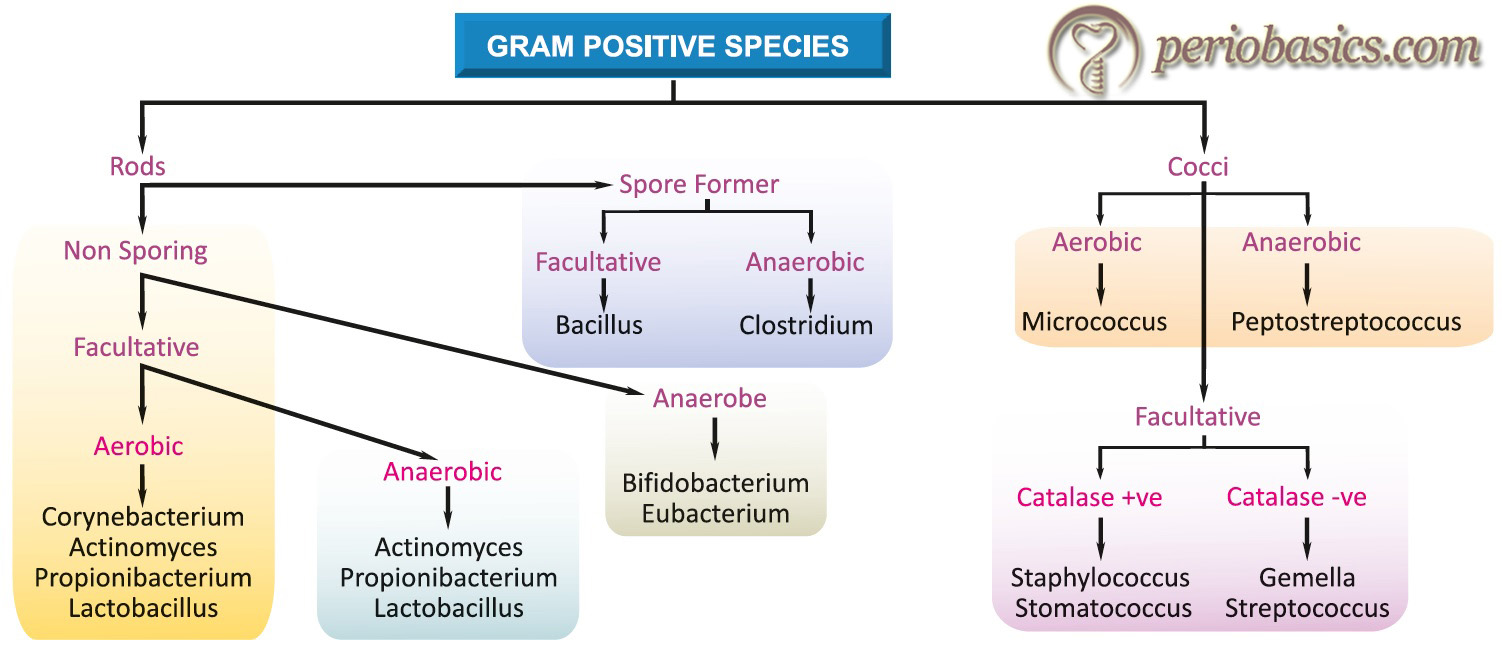
Systematic classification and categorization of organisms into ordered groups is called taxonomy. A working knowledge of bacterial taxonomy is essential for understanding various bacterial species involved in the periodontal disease progression. Although higher organisms are classified according to their evolutionary pathways (i.e. phylogenetically), bacteria cannot be similarly categorized because of the insufficiency in their morphological features. Bacterial classification is somewhat artificial in that they are categorized according to phenotypic features, which facilitate their laboratory identification. These comprise
- Morphology (cocci, bacilli, spirochetes etc.).
- Staining properties (Gram-positive, Gram-negative etc.)
- Spore formers or non-spore formers.
- Cultural requirements (aerobic, facultative anaerobic, anaerobic etc.).
- Biochemical reactions (saccharolytic and asaccharolytic according to sugar fermentation reaction).
- Antigenic structure (serotypes).
Bacterial classification according to the above phenotypic features has been illustrated in the following figures, where bacterial species have been divided into Gram-positive and Gram-negative as per their staining properties.

Establishing an organism as a periodontal pathogen
Periodontal diseases are caused by a variety of micro-organisms that reside at or below the gingival margin in the form of plaque biofilm. Because there is a complex microbiota involved in the formation of plaque biofilm, first we need to distinguish the pathogenic bacterial species from the host-compatible species. Secondly, we need to devise treatment methods that can decrease the pathogens while maintaining host-compatible species. But, how to establish the association of a microorganism with the pathogenesis of a particular periodontal disease?
To establish an association of an organism with disease pathogenesis, three rules for experimental proof for the pathogenicity of an organism were presented in 1883 by the German bacteriologist, Robert Koch, and a fourth rule was appended by Smith (1905) 10. These rules of proof are often referred to as Koch’s Postulates. These are,
1. The suspected causal organism must be constantly associated with the disease.
2. The suspected causal organism must be isolated from an infected host and grown in pure culture.
3. When a healthy susceptible host is inoculated with the pathogen from pure culture, symptoms of the original disease must develop.
4. The same pathogen must be re-isolated from the host infected under experimental conditions.
However, periodontal diseases are not caused by a single organism which is the cornerstone of Koch’s postulates. Indeed, these disease entities are the result of mixed infections. To better identify periodontopathogenic bacterial candidates Koch’s postulates were replaced with Socransky’s postulates 11. These include the following:
1. The organism must be found in relatively high numbers in proximity to the periodontal lesion.
2. The organism must be absent, or present in much smaller numbers in periodontally healthy subjects or in subjects with other forms of periodontal disease.
3. The organism must have high levels of serum, salivary and gingival crevicular fluid antibody developed against it in periodontally diseased subjects.
4. The organism must be found to produce virulence factors in vitro, which can be correlated with clinical histopathology.
5. The organism must mimic similar pathogenic properties in an appropriate animal model.
6. Clinical improvement following treatment must eliminate the putative pathogen from the periodontal lesion.
Various microorganisms are involved in the pathogenesis of periodontal diseases. But along with them, many micro-organisms are …… Contents available in the book…. Contents available in the book…… Contents available in the book……
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.
India Users:
International Users:
Subgingival microbial complexes
In a landmark study, Socransky et al. (1998) 13 examined over 13,000 subgingival plaque samples from 185 adult subjects and used DNA hybridization methodology and community ordination techniques to demonstrate the presence of specific microbial groups within the dental plaque. The presence and levels of 40 subgingival taxa were determined in plaque samples using whole genomic DNA probes and checkerboard DNA-DNA hybridization. According to the presence of microorganisms and the related periodontal status, these organisms were grouped into five complexes,
- 1st complex (Red complex): Tannerella forsythia, Porphyromonas gingivalis, and Treponema denticola.
- 2nd complex (Orange complex): Eubacterium nodatum, Campylobacter rectus, Campylobacter showae, Streptococcus constellatus, and Campylobacter gracilis.
- 3rd complex (Yellow complex): Streptococcus sanguis, S. oralis, S. mitis, S. gordonii, and S. intermedius.
- 4th complex (Green complex): Campylobacter concisus, Eikenella corrodens, and Actinobacillus actinomycetemcomitans serotype a.
- 5th complex (purple complex): Veillonella parvula and Actinomyces odontolyticus.
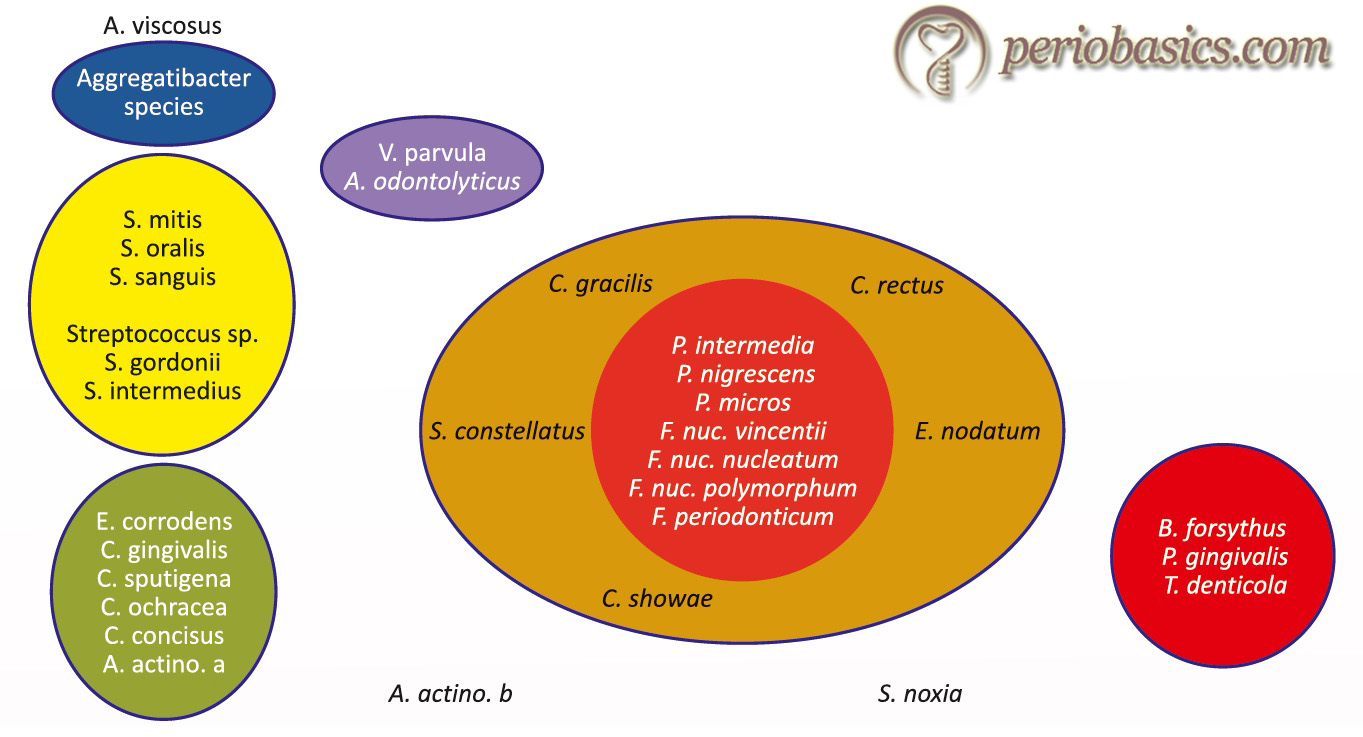
A. actinomycetemcomitans serotype b, Selenomonas noxia and Actinomyces naeslundii genospecies 2 (A. viscosus) were outliers with little relation to each other and the 5 major complexes.
Out of these groups of species the Actinomyces species, yellow, green and purple complexes are early colonizers of the tooth surface whose growth usually precedes the multi-plication of the predominantly Gram-negative orange and red complexes. The orange and red complexes are comprised of the species thought to be the major etiologic agents of periodontal diseases.
Microorganisms associated with periodontal health
As discussed in “Dental plaque”, plaque formation starts soon after cleaning the tooth surface and the early dental plaque has been shown to be dominated (60%-90%) by Streptococcus species. Other bacterial species found in early plaque include Eikenella species, Haemophilus species, some Prevotella species, Capnocytophaga species, Propionibacterium species, and Veillonella species 14. It has been shown that Actinomyces species is the predominant species after 2 hours of biofilm formation in healthy subjects 15. As the maturation of plaque takes place, microorganisms also called ‘tertiary colonizers’ can be isolated from the plaque samples. These include A. actinomycetemcomitans, P. intermedia, Eubacterium species, Treponema species, and Porphyromonas gingivalis. F. nucleatum serves to bridge between the early and late colonizers.
Typically, healthy gingival sites have a bacterial pattern which is similar to, as described for the relatively immature supragingival plaque. The majority of the bacterial counts from plaque samples from healthy sites consistently have bacterial species that belong to Streptococci, Actinomyces species (especially, A. viscosus and A. naeslundii) and Veillonella species 16-18. It has been demonstrated that if a plaque sample is taken from healthy periodontal sites and is examined in a wet mount using phase-contrast or darkfield microscopy, the bacteria seen are primarily non-motile with a ratio of motile to non-motile forms of about 1:40 19. This study also provides evidence that in diseased periodontal sites the number of motile forms of bacteria increases. There are many other bacterial species that can be …… Contents available in the book…. Contents available in the book…… Contents available in the book……
| Periodontal Condition | Associated micro-organisms |
|---|---|
| Periodontal Health | Streptococcus oralis, Streptococcus sanguis, Streptococcus mitis, Actinomyces gerencseriae, Actinomyces naeslundii, Fusobacterium species, Prevotella nigrescens, Veillonella species. |
| Gingivitis | Lactobacillus species, Actinomyces naeslundii, Peprostreptococcus micros, Streptococcus anginosus, Fusobacterium nucleatum, P. intermedia, Winonalla parvula, Campylobacter species, Haemophilus species, Selenomonas Species, Treponema species. |
| Periodontitis with slow/moderate rate of progression (Grade A/B) | Eubacterium brachy, Eubacterium nodatum, Mogibacterium timidium, Parvimonas micra, Peptostreptococcus stomatis, Parvimonas micra, Tannerella forsythia, Fusobacterium nucleatum, Porphyromonas gingivalis, Prevotella intermedia, Prevotella loescheii, Dialister pneumosintes, Campylobacter rectus, Treponema species. |
| Periodontitis with rapid rate of progression (Grade C) | Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, Prevotella intermedia, Prevotella nigrescens, Eikenella corrodens, Selenomonas sputigena, F. nucleatum, Campylobacter rectus, Peptostreptococcus micros, Campylobacter concisus. |
| Refractory periodontitis | Aggregatibacter actinomycetemcomitans, Streptococcus constellatus/intermedius, Tannerella forsythia, P. gingivalis, T. denticola, Campylobacter rectus, Eikenella corrodens. |
| Necrotizing ulcerative gingivitis/periodontitis | Prevotella intermedius, Treponema species, Selenomona species, Fusobacterium species, Candida species. |
Microorganisms associated with periodontal diseases
Specific microorganisms have been implicated in various periodontal conditions by various studies. However, it must be remembered that because periodontal diseases are primarily the result of complex microbial infection, the majority of these organisms are common in various periodontal conditions. Attempts have been made to make some general comparisons of the subgingival microbial profiles associated with chronic and aggressive forms of periodontitis. Mombelli et al. (2002) 20 in a systemic review, tried to make a comparison between chronic and aggressive periodontitis on the basis of presence or absence of most commonly identified putative periodontal pathogens P. gingivalis, A. actinomycetemcomitans, P. intermedia, T. forsythia and C. rectus in patients with chronic and aggressive periodontitis. The results of the study could not conclude any relation of any of these microorganisms specifically to chronic or aggressive periodontitis. However, commonly identified microorganisms in gingivitis, Grade A/B (chronic) periodontitis, and Grade C (aggressive) periodontitis can be listed based on extensive research work done on the microbiology of various periodontal conditions.
Microorganisms associated with gingivitis
The early studies on gingivitis demonstrated that with the development of gingivitis, there is a gradual microbial shift from a Gram-positive dominated microflora to more complex flora that contains significantly higher numbers of Gram-negative and spiral forms of bacterial species 21. Later studies, on the development of gingivitis, elaborated that the bacterial culture of plaque gradually shifts from predominantly Strepto-coccus to that dominated by Aggregatibacter species 17, 22. Furthermore, it was also observed that although the number of Aggregatibacter species increases with the increase in plaque mass, but bleeding on probing was associated with increased numbers of A. viscosus and pigmented Bacteroides. More extensive studies on gingivitis have demonstrated that sites with gingivitis have increased numbers of F. nucleatum, Eubacterium timidum, Treponema species and Bacteroides species 23. Most of the bacterial species isolated from gingivitis associated plaque primarily belong to Aggregatibacter, purple and yellow complexes with partial representation from the orange complex.
Periodontitis with slow to moderate rate of progression (Grade A and B)
Extensive research has been done to identify the microbiota associated with periodontitis with a slow to moderate rate of progression (chronic periodontitis). The untreated chronic periodontitis patients have a predictable pattern of subgingival plaque maturation. Although the composition of microbiota may vary a little depending on how the ecosystem during plaque maturation has been disrupted by oral hygiene methods, most of the patients with chronic periodontitis demonstrate almost consistent bacterial species in the subgingival plaque samples. The most consistent micro-organisms isolated from chronic periodontitis cases include P. gingivalis and T. forsythia 24. Other organisms that have been shown to be associated with chronic periodontitis include F. nucleatum, P. intermedia, Veillonella parvula, Campylobacter species, Haemophilus species, Selenomonas species, and Treponema species.
The recent non-culture based identification techniques have identified microorganisms from Archaea domain in association with periodontal diseases. These microorganisms are prokaryotes that physically resemble bacteria, but have different nucleotide sequences, hence cannot be considered in the bacteria domain. Instead, they are genetically closer to microbes in the Eukarya domain. In chronic periodontitis cases, approximately 19-73 % of subgingival sites have been shown to harbor Archaea 25-28. It must be noted that Archaea have no direct pathogenic effects, but these can contribute to the overall pathogenicity of subgingival biofilms by various syntrophic interactions with other microorganisms present in the plaque 29.
Another organism that has been identified with non-culture techniques is Filifactor alocis 30. This microorganism is a fastidious, Gram-positive, obligately anaerobic rod which was repeatedly identified in periodontal lesions, using DNA-based methods. It has been suggested to be a marker for periodontal deterioration. In a study, plaque samples derived from healthy, chronic periodontitis and aggressive periodontitis patients were subjected to PCR analysis. Results showed that patients suffering from generalized aggressive periodontitis or chronic periodontitis harbored F. alocis whereas it was rarely detected in the control group. The authors concluded that F. alocis is a contributor to the pathogenetic structure of biofilms accounting for periodontal inflammation and can be considered as an excellent marker microorganism for periodontal disease 30.
Periodontitis with a rapid rate of progression (Grade C)
The localized and generalized forms of periodontitis with a rapid rate of progression (aggressive periodontitis) result in rapid periodontal destruction. The most commonly implicated microorganism in aggressive periodontitis is Aggregatibacter actinomycetemcomitans 31, 32. This microorganism is often found in high numbers in sites with aggressive periodontal destruction and is rarely seen in healthy subjects, those with plaque-induced gingivitis, or edentulous patients 33-35. Other microorganisms that are routinely isolated from aggressive periodontitis cases include Capnocytophaga species and Prevotella species. Studies have shown that infection with A. actinomycetemcomitans is not substantially reduced following the non-surgical periodontal therapy 36, 37, but may be reduced by systemic antibiotic and surgical therapy 35, 37, 38. These findings indicate that A. actinomycetemcomitans …… Contents available in the book…. Contents available in the book…… Contents available in the book……
Refractory periodontitis
According to the American Academy of Periodontology (AAP) 44, refractory periodontitis is not a single disease entity. The term refers to destructive periodontal diseases in patients who, when longitudinally monitored, demonstrate additional attachment loss at one or more sites, despite well-executed therapeutic and patient efforts to stop the progression of the disease. It has been reported that microbiota of the refractory periodontitis, on an average, is generally similar to that observed in chronic periodontitis 45. However, some differences have been pointed out. Magnusson et al. (1991) 46 and Colombo et al. (1998b) 45 have pointed out high levels of Streptococcus constellatus/intermedius in refractory periodontitis lesions while Gordon et al. (1985) 47 have observed high levels of motile organisms and black pigmented species in these cases. High levels of Aggregatibacter actinomycetemcomitans were observed by van Winkelhoff et al. (1992) 48 in refractory periodontitis cases. Winkel et al. (1997) 49 found high levels of T. forsythia in these cases. Haffajee et al. (1988) 50 in a study on 13 refractory periodontitis subjects found that the microbial profile of these patients, primarily contained members of the red and/or orange complexes. Thus, it can be summarized that the microbial profile of refractory periodontitis patients is not very different from that of chronic and aggressive periodontitis. However, according to the specifically identified microorganisms, individual treatments may have to be designed for every patient.
Necrotizing ulcerative gingivitis and periodontitis
Necrotizing periodontal diseases are the result of microbial infection in association with attenuated immune response commonly due to malnutrition, stress or HIV infection. There are specific microorganisms that have been isolated from necrotizing periodontal lesions. These include Prevotella intermedia and Treponema species (intermediate-sized oral treponemes), Selenomonas species and Fusobacterium species 51, 52. The microorganisms isolated from HIV-associated periodontitis have been found to be similar to that observed in conventional periodontitis patients 53-55. Furthermore, Candida albicans infection has also been associated with necrotizing ulcerative gingivitis in HIV seropositive patients 56.
Factors that determine the initiation and progression of the periodontal disease
There are many factors that determine the establishment and proliferation of microorganisms in a host. These can be broadly divided into pathogen and host-related factors. The pathogen should have properties which enable it to overcome the host immune response and to adhere and proliferate on or in the host. The host-related factors include the defense mechanism which prevents the pathogen to establish and proliferate in the host. These factors are affected by the local and environmental factors which may or may not favor the establishment of infection. Let us now discuss pathogen virulence, host susceptibility and other factors which affect the occurrence of a disease.
Virulence of microorganisms
Virulence factors refer to the properties that enable a microorganism to establish itself on or within a host of a particular species and enhance its potential to cause disease. These factors, primarily perform three functions,
- Invade the host.
- Evade host defenses.
- Cause disease.
These factors include cell-surface proteins that mediate the bacterial attachment, cell surface carbohydrates, and proteins that protect a microorganism, bacterial toxins and hydrolytic enzymes that may contribute to the pathogenicity of the bacterium. Bacterial adherence to the host is the first step in the development of the disease. Many pathogenic bacteria colonize mucosal sites by using pili (fimbriae) to adhere to the cells. In the context of periodontal disease progression, initial adherence, colonization and growth of microorganisms in dental biofilm is the first step in disease progression. The attachment of bacteria to dental pellicle exhibits a great deal of specificity and appears to involve specific receptors on the bacterial and pellicle surfaces. Plaque accumulation may be mediated by bacterial extracellular polysaccharides, salivary components as well as direct cell-to-cell binding.
Salivary components play a dual role in plaque formation. They can mediate bacterial attachment to the pellicle or plaque periphery, but by binding to unattached bacteria they can also diminish the attachment of such bacteria. As the plaque matures, bacterial coaggregation can be seen. Coaggregation prevalent among bacteria isolated from the human oral cavity was first reported by Gibbons and Nygaard in 1970 57. Once established in a biofilm, these factors start producing their respective virulence factors which trigger the host immune response. It must be noted that different strains of a species vary in their virulence. Certain clonal types of a particular bacterial species are more commonly found in subjects with periodontitis as compared to periodontally healthy patients. It has been demonstrated in animal experiments that the virulence of different strains varies for P. gingivalis 58-61.
To cause disease, a pathogen should have all the necessary genetic information regarding virulence factors which may enable the establishment of that pathogen in the host. If some of the elements of this genetic information are lacking in a pathogen, these could be received from other strains of that species (or possibly other species) via phage, plasmids or transposons 62. Finally, the microorganism should be located at a right location in sufficient numbers (which varies from species to species) to initiate the disease process.
Many bacterial species are equipped with various virulence factors to overcome the host defense mechanisms. These virulence factors vary amongst bacterial species and within a species from strain to strain. Certain toxins such as leukotoxins cause damage to polymorphonuclear leukocytes (PMN’s), which make the first line of defense against bacterial invasion in the tissue. For example, leukotoxin by A. actinomycetemcomitans destroys leukocytes, thus facilitating bacterial invasion 63. These leukotoxins have also been found to be effective against monocytes 63 and mature T- and B-lymphocyte cell lines 64. Furthermore, specific mechanisms that interfere with killing mechanisms of PMN’s have been developed by various bacterial species 65-67. Anti-phagocytosis factors such as bacterial capsule prevent the bacterial phagocytosis by macrophages. Specific antibody response (IgA and IgG) has been detected against …… Contents available in the book…. Contents available in the book…… Contents available in the book……
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.
India Users:
International Users:
To effectively evade the host defense system, a microorganism should have sophisticated mechanisms to prevent its inactivation or killing. Complex mechanisms have been explained for various microorganisms which modulate or suppress the immune response, thus ensuring bacterial survival 71, 72. Once, a periodontal pathogen overcomes the host immune response; it may cause tissue damage by producing various toxins and enzymes that degrade the components of the connective tissue.
Host susceptibility
A susceptible host is the one who responds to the infection in a different way as compared to the normal individuals. Recent research has enumerated various “host susceptibility factors” based on various new methods for comparing the immunological responses in subjects with periodontitis and healthy individuals. These factors include poorly regulated immune response and defective PMN function. Systemic conditions like diabetes mellitus, HIV infection, etc. have been shown to have a direct relation to periodontal disease progression.
Local factors
The local environment in the subgingival area is a major factor that determines the establishment and survival of various bacterial species. Although, mostly the subgingival environment is host-compatible and favors the growth of commensals, sometimes due to microorganism related or host-related factors, the subgingival environment becomes conducive for the growth of putative periodontal pathogens.
Transmission of bacterial pathogens
It is a well-known fact that development of the disease can be prevented if the microorganism causing that disease can be prevented from entering the body. Thus, if we can prevent the entry of periodontal pathogens in the oral cavity, the progression of the periodontal disease process can be prevented. But from where these microorganisms reach the oral cavity? The most commonly identified periodontal pathogens are not usually found in our surrounding environments such as soil, water, and air. Hence, most probably, these microorganisms are transmitted from one person to another. The transmission of the microorganisms can be horizontal or vertical. The vertical transmission is from parents to offspring. Any mode of transmission other than the parent-offspring transmission is referred to as horizontal transmission.
The epidemiological studies based on molecular identification techniques have provided us with the evidence regarding the transmission of periodontal pathogens in a vertical and horizontal manner. The basic principle on which these studies are based is the isolation of the bacterial strain, extraction of its DNA and cutting this DNA with restriction endonucleases. It is then run on agarose gel electrophoresis, which provides us the fingerprint of the DNA. This fingerprint pattern of DNA is then compared with already known DNA fingerprints. It was observed that A. actinomycetemcomitans and P. gingivalis strains isolated from parents and children from the same family had an identical restriction endonuclease pattern 73-76. Studies done to identify horizontal transmission have found similar A. actinomycetemcomitans and P. gingivalis strains isolated from husband and wife demonstrating that the periodontal pathogens can be transmitted within the married couples 77, 78. However, it must be noted that the transmission of these bacterial species is not just limited to family members. JP2 clone of A. actinomycetemcomitans has been found in individuals of African descent who relocated from Africa to Europe or America 79.
Hence, different clonal types of the same species are introduced in an individual throughout his or her lifetime. As some clonal types of a bacterial species are more virulent than the other clonal types, the pattern of disease progression changes with the introduction of more virulent clonal type in an individual. The transmission of bacterial species may also occur between different biofilms. Such as, …… Contents available in the book…. Contents available in the book…… Contents available in the book……
Periobasics: A Textbook of Periodontics and Implantology
The book is usually delivered within one week anywhere in India and within three weeks anywhere throughout the world.
India Users:
International Users:
Conclusion
Micro-organisms are the primary etiology of periodontal diseases. Our oral cavity has a variety of micro-organisms. Most of them are commensals and live in harmony with us without causing any disease. However, in certain situations, they may become pathogenic and cause disease. The accumulation and proliferation of various pathogenic microbial species in the periodontal arena in susceptible individuals may cause periodontitis. In the next articles, we will read about the major periodontal pathogens.
References
References are available in the hard-copy of the website.